Appendix A MLTD 2203 Clinical Microbiology IV Test Procedures
Anaerobe Identification Disks
Remel Anaerobe Identification Disks
Special potency identification discs are used to presumptively identify anaerobic Gram-negative bacilli. Colistin and vancomycin confirm the Gram stain reactions. Penicillin, rifampin and kanamycin are used to separate Bacteroides spp. and Fusobacterium spp.
- Allow discs to warm to room temperature.
- Select one well-isolated colony and streak the first quadrant of the plate back and forth several times to ensure an even lawn of growth. Streak the remaining quadrants for isolation.
- Using flamed forceps, place the colistin, kanamycin and vancomycin discs in the first quadrant, well separated from each other. Subsequent discs can be placed in the second quadrant.
- Aerotolerance testing of the test isolate should be performed at this time if not already established.
- Incubate plate anaerobically at 35-37°C for 24-48 hours or longer until adequate growth is evident. Measure zone of inhibition and record.
Interpretation
- Sensitive = zone of inhibition greater than 10 mm
- Resistant = No zone of inhibition or zone less than 10 mm
Expected values
| VAN | KAN | COL | PEN | RIF | |
|---|---|---|---|---|---|
| Gram positive | S | V | R | – | – |
| Gram negative | R | R | S | – | – |
| Bacteroides fragilis group | R | R | R | R | – |
| Other Bacteroides spp. | R | R | V | S | – |
| Pigmented Porphyromonas spp. | S | R | R | – | – |
| Fusobacterium spp. | R | S | S | – | Varies with species |
| Bacteroides ureolyticus group | R | S | S | – | – |
API® 20 E TEST PROCEDURE
Biomerieux® api® 20 E Procedure
For use with Enterobacteriaceae and/or non-fastidious Gram-negative rods.
- Label the elongated flap on the incubation tray with the specimen identification, the date and your initials.
- Dispense about 5-8 mL of distilled water into the incubation tray to provide humidity during incubation. An accurate volume is not critical; just be sure that all the small wells are filled with water.
- Record your specimen identification on an api® worksheet in the box labelled “REF”. Also record your initials and the date and time that you set up the test.
- Label a 5-mL tube of sterile saline (api® NaCl 0.85% Medium or homemade) with the specimen identification.
- Use a sterile stick, loop or swab to remove an entire well-isolated colony from an 18-24 culture grown on a medium that is appropriate for growth of Enterobacteriaceae and non- fastidious Gram-negative bacilli. Carefully emulsify the entire colony in the suspension. The saline should appear slightly cloudy.
- Remove an api® 20E strip from its envelope and place it in the incubation tray. Note: you may place the incubation lid under one long edge of the tray to tilt the strip on an angle. This may make inoculation of the strip easier.
- Use a sterile Pasteur pipette to gently aspirate a portion of the organism suspension from the saline vial. Using aseptic technique, place the pipette tip against the inner side of each cupule in the strip and fill the tube section gently. Proceed slowly and do your best to avoid bubbles. If bubbles do occur, finish filling all of the tubes and then use a loop, straight wire or sterile stick to tease out the bubbles. If you are removing bubbles from more than one tube, flame the loop or replace the stick between each one to avoid carryover.
- Fill the tube and cupule sections of the Citrate (CIT), VP and GEL tubes. Fill only the tube portions of the other tests.
- After inoculation, fill the cupules of the following tests with sterile mineral oil: ADH, LDC, ODC, H2S, URE
- Place the plastic lid on the incubation tray and incubate in ambient air at 35-37°C for 18-24 hours.
- Inoculate 1 drop of the saline suspension to a labelled blood agar purity plate. Streak for isolated colonies. Incubate in ambient air at 35-37°C for 18-24 hours. Discard the saline suspension in biohazard waste.
- Perform an oxidase test on an isolated colony of your unknown organism. Only perform this test on colonies from a non-selective, non-differential medium. Record the result in the appropriate space on the api® worksheet.
api® 20E Results
- After incubation, record all results on the api® worksheet.
- First check your purity plate to ensure that there has been no contamination of your organism suspension. Do this by comparing the colony morphology to the original colony description from the previous day and ensuring that it matches. Record the purity as “OK” or “acceptable”.
- If 3 or more tests are positive on the api® strip, proceed with reading all of the spontaneous tests. If there are less than 3 positive tests (including GLU), re-incubate the strip for an additional 24 hours without adding reagents.
- Read the spontaneous tests
Positive Negative ONPG yellow (even if pale yellow) colorless. Compare the color to the VP tube (before reagents have been added) if in doubt. ADH, LDC, ODC (decarboxylase/dihydrolysis reactions) red or orange yellow (If orange after 36-48 hours incubation →neg) CIT blue or blue-green negative pale green or yellow *Read in the cupule (aerobic portion of tube) H2S black deposit/thin line colorless/grey URE red or orange yellow. Skip the IND, TDA and VP tests for now.
GEL diffusion of black pigment throughout the tube no diffusion GLU yellow or greyish yellow blue or blue-green Other carbohydrate fermentation tests (MAN → ARA) yellow blue or blue-green Note for all carbohydrates: fermentation begins in the lower portion of the tube. Oxidation begins in the cupule. - Add one drop of ferric chloride to the TDA tube: pos = reddish brownNeg = yellow (or colour of reagent)
- Add one drop of 40% KOH and one drop of alpha-naphthol to the VP tube. Incubate at room temperature for 10 minutes.
- Pos = pink/red
- Neg = colourless/pale pink
- Add one drop of Kovac’s reagent or James reagent to the IND tube.
- pos = pink
- neg = colorless/pale green or yellow
- On the api® worksheet, determine the numerical profile: a number has been assigned to each test in the strip. The tests are divided into groups of three. For each test that is positive, add up the assigned numbers and record the total of the three tests on the square or circle below. In this way, a seven-digit number will be produced. To determine the most probable identity of your organism, refer to the api® website.
- From the website, record the most likely organism identification on your worksheet. The identification must be described as “excellent”, “very good” or “good” for it to be acceptable. Record this comment on your worksheet. The percentage of likelihood that is indicated should also be recorded. Generally, a percentage of greater than 90% is required for an acceptable identification. If the identification is described as “low selectivity”, “low discrimination”, “doubtful profile” or “unacceptable ID”, re-check/re- read your reactions and the addition of the profile numbers. If an error was made originally, go back to the api® website and type in your new profile number.
- See the package insert for supplementary tests. These may be required if the 7-digit profile is not discriminatory enough. The wet motility (MOB) and glucose OF tests are performed externally. Record the MacConkey test as “positive” if the organism grows on MacConkey agar and “negative” if it does not.
- For the supplementary nitrate reduction test, add 1 drop of NIT 1 and 1 drop of NIT 2 to the GLU tube after the GLU result has been recorded. Wait 2-5 minutes.
- Pos = red (record as “pos” in the NO2 bubble on the api worksheet)
- Neg = yellow
To determine if nitrite has been produced and then reduced to nitrogen gas, add 2-3 mg of zinc to the GLU tube. After 5 minutes, record “+” for N2 if the tube remains yellow.
Record a negative result for N2 if the tube turns orange-red.
Notes
- Generally, api® reactions are more sensitive than conventional tube media except for the urea test. If the api® urea result is negative and a tube urea test is positive on the same organism, the result from the tube urea can be used in determining the organism profile number. Just be sure to make a note on the worksheet that this is where the result was obtained from.
- If Salmonella or Shigella are identified by api®, serological tests must be performed to confirm their identity.
api® Candida Test Procedure
The API® (Analytical Profile Index) Candida system (Biomerieux®) is a rapid, computer-based micro method for the separation and identification of yeasts frequently encountered in clinical microbiology. The system consists of 10 micro cupules containing dehydrated substrates for the performance of 12 sugar acidification or enzymatic identification tests. The addition of suspension of the test organism serves to hydrate the media and to initiate the biochemical reactions. The reactions produced during incubation then result in spontaneous color changes. The identification of the unknown yeast species is made by comparing the results with a list of profiles in the identification software.
- Label the elongated flap on the incubation tray with the specimen identification, the date and your initials.
- Dispense about 5-8 mL of distilled, deionized water into the incubation tray to provide humidity during incubation. An accurate volume is not critical; just be sure that all of the small wells are filled with water.
- Record the specimen identification on an api® Candida worksheet in the box labelled “REF”. Also record your initials and the date and time that you set up the test.
- The colonies for testing should be 18-24 hours old and grown on blood agar, Albicans ID 2 agar or Sabouraud 2 agar plates. Ensure that you have pure colonies and that the organism appears yeast-like microscopically.
- Label an ampoule of api® 0.85% NaCl medium (2 mL) with the specimen identification and open it carefully.
- Use a sterile stick or swab to prepare a homogeneous bacterial suspension with a turbidity equivalent to 3 McFarland. Use the suspension to set up the strip immediately after preparation.
- Remove an api® Candida strip from its envelope and place it in the incubation tray. Note: you may place the incubation lid under one long edge of the tray to tilt the strip on an angle. This may make inoculation of the strip easier.
- Use a sterile Pasteur pipette to gently aspirate a portion of the organism suspension from the vial containing the inoculated NaCl medium. Using aseptic technique, place the pipette tip against the inner side of each cupule in the strip and fill each tube section gently. Proceed slowly and do your best to avoid bubbles. If bubbles do occur, finish filling all of the tubes and then use a loop, straight wire or sterile stick to tease out the bubbles. If you are removing bubbles from more than one tube, flame the loop or replace the stick between each one to avoid carryover.
- Cover the first five tests (GLU to RAF) and the last test (URE) with mineral oil immediately after inoculating the strip.
- Place the plastic lid on the incubation tray and incubate in ambient air at 34-38°C for 18- 24 hours.
After incubation
- Check your purity plate to ensure that there has been no contamination of your organism suspension. Do this by comparing the colony morphology to the original colony description from the previous day and ensuring that it matches. Record the purity as “OK” or “acceptable”.
- Record all results on the api® worksheet.
Summary Positive Negative Carbohydrate acidification tests (GLU→RAF) yellow or green/gray violet or grey violet βMA pale yellow to bright yellow colorless αAMY pale yellow to bright yellow colorless βXYL pale yellow to bright yellow colorless to very pale yellow/ blue/green** (Note: any trace of green in cupule 8= βXYL neg/ βNAG pos)
βGUR pale yellow to bright yellow colorless/blue/green URE red yellow-pale orange βNAG (in tube #8) blue/green** colorless/yellow (Note: any trace of green in cupule 8= βXYL neg/ βNAG pos) βGA (in tube #9) blue/green colorless/yellow
- On the api® worksheet, determine the numerical profile: A number has been assigned to each test in the strip. The tests are divided into groups of three. For each test that is positive, add up the assigned numbers and record the total of the three tests on the square or circle below. In this way, a seven-digit number will be produced. To determine the most probable identity of your organism, refer to the api® website.
- From the website, record the most likely organism identification on your worksheet. The identification must be described as “excellent”, “very good” or “good” for it to be acceptable. Record this comment on your worksheet. The percentage of likelihood that is indicated should also be recorded. Generally, a percentage of greater than 90% is required for an acceptable identification. If the identification is described as “low selectivity”, “low discrimination” or “unacceptable ID”, re-check/re-read your reactions and the addition of the profile numbers. If an error was made originally, go back to the api® website and type in your new profile number.
API® STAPH TEST PROCEDURE
The API® (Analytical Profile Index) STAPH ® system (developed by Analytab Products, Division of Sherwood Medical, Plainview, New York) is a rapid, computer-based micro method for the separation and identification of the 13 species of staphylococci. The system consists of 10 micro cupules containing dehydrated substrates for the performance of conventional and chromogenic tests. The addition of suspension of the test organism serves to hydrate the media and to initiate the biochemical reactions. The identification of the staphylococcal species is made with the aid of differential charts or the STAPH Profile Register that is part of the system or both.
Biomerieux® api® Staph Procedure: For use with Gram positive cocci that are catalase positive,
e.g. Staphylococcus, Micrococcus and Kocuria spp.
- Label the elongated flap on the incubation tray with the specimen identification, the date and your initials.
- Dispense about 5-8 mL of distilled, deionized water into the incubation tray to provide humidity during incubation. An accurate volume is not critical; just be sure that all of the small wells are filled with water.
- Record the specimen identification on an api® Staph worksheet in the box labelled “REF”. Also record your initials and the date and time that you set up the test.
- The colonies for testing should be 18-24 hours old and grown on Columbia agar plates. Ensure that you have pure colonies and that the organism is a Gram-positive coccus that is catalase positive
- Label an ampoule of api® Staph medium with the specimen identification and open it carefully.
- Use a sterile or swab to prepare a homogeneous bacterial suspension with a turbidity equivalent to 0.5 McFarlane. Use the suspension to set up the strip immediately after preparation.
- Remove an api® Staph strip from its envelope and place it in the incubation tray. Note: you may place the incubation lid under one long edge of the tray to tilt the strip on an angle. This may make inoculation of the strip easier.
- Use a sterile Pasteur pipette to gently aspirate a portion of the organism suspension from the vial containing the inoculated Staph medium. Using aseptic technique, place the pipette tip against the inner side of each cupule in the strip and fill the tube section gently. Proceed slowly and do your best to avoid bubbles. If bubbles do occur, finish filling all of the tubes and then use a loop, straight wire or sterile stick to tease out the bubbles. If you are removing bubbles from more than one tube, flame the loop or replace the stick between each one to avoid carryover.
- After inoculation, fill the cupules of the following tests with sterile mineral oil: ADH and URE
- Note: this ensures anaerobiosis
- Place the plastic lid on the incubation tray and incubate in ambient air at 35-37°C for 18-24 hours.
- Inoculate 1 drop of the saline suspension to a labelled blood agar purity plate. Streak for isolated colonies. Incubate in ambient air at 35-37°C for 18-24 hours. Discard the Staph medium suspension in biohazard waste.
After incubation
- Record all results on the api® worksheet.
- Check your purity plate to ensure that there has been no contamination of your organism suspension. Do this by comparing the colony morphology to the original colony description from the previous day and ensuring that it matches. Record the purity as “OK” or “acceptable”.
- Develop the reactions as follows:
-
- VP test: Add I drop pf VP1 reagent + 1 drop of VP2 Wait 10 minutes. A violet-pink color indicates a positive reaction. Pale pink or light pink is negative.
- NIT test: Add 1 drop of NIT1 reagent + 1 drop of NIT2 Wait 10 minutes. A red color indicates a positive reaction.
- PAL test: Add 1 drop of ZYM A reagent + 1 drop of ZYM b Wait 10 minutes. A violet color indicates a positive reaction.
Summary Positive Negative NIT red colorless to light pink PAL violet yellow VP violet-pink colorless to light pink
-
- While the NIT, PAL and VP tests are developing, read and record the spontaneous test results: 0: negative = red. This well has no substrate. If the color is not red at the end of the incubation, the GLU→MEL tests are not valid.
Positive Negative 0 red. This well has no substrate. If the color is not red at the end of the incubation, the GLU→MEL tests are not valid.
GLU, FRU, MNE, MAL, LAC, TRE, MAN, XLT, MEL (carbohydrate fermentation) yellow red (If MNE and XLT are orange and tests preceding or following them are positive, consider them to be negative). RAF, XYL, SAC, MDG, NAG yellow red ADH orange-red yellow URE red-violet yellow - Obtain the lysostaphin resistance test result from the instructor and record it as the 21st test result on the worksheet.
- On the api® worksheet, determine the numerical profile: A number has been assigned to each test in the strip. The tests are divided into groups of three. For each test that is positive, add up the assigned numbers and record the total of the three tests on the square or circle below. In this way, a seven-digit number will be produced. To determine the most probable identity of your organism, refer to the api® website.
- From the website, record the most likely organism identification on your worksheet. The identification must be described as “excellent”, “very good” or “good” for it to be acceptable. Record this comment on your worksheet. The percentage of likelihood that is indicated should also be recorded. Generally, a percentage of greater than 90% is required for an acceptable identification. If the identification is described as “low selectivity”, “low discrimination” or “unacceptable ID”, re-check/re-read your reactions and the addition of the profile numbers. If an error was made originally, go back to the api® website and type in your new profile number.
Bacitracin Procedure
BBL™ Bacitracin Test
- Use a sterile loop or swab to streak an isolated colony of a beta haemolytic Streptococcus to a trypticase soy agar with 5% sheep blood (BA plate).
- Use heated forceps to apply a 0.04 U bacitracin disk to the well or first quadrant (area of heaviest growth) of the plate.
- Incubate the plate at 35-37°C for 18-24 hours in ambient air for staphylococci or 5-10% CO2 for streptococci.
- Look for an area of inhibition around the disk.
Pos = area of inhibition (Presumptive ID of Streptococcus pyogenes)
Neg = no area of inhibition.
Positive QC: Streptococcus pyogenes
Negative QC: other beta haemolytic streptococci (e.g. Streptococcus agalactiae)
Note: Groups C and G streptococci are also susceptible to bacitracin so this test should only be used to identify Group A Streptococcus if it is done in conjunction with a PYR test.
Bacticard® Candida
Remel Bacticard® Candida
This tests aids in the identification of Candida albicans by the rapid detection of L-proline aminopeptidase and β-galactosaminidase.
- Label Bacticard® Candida with the specimen identification.
- Add one (1) drop of Bacticard® Candida Rehydrating Fluid to each test circle. Do not oversaturate the test area.
- Inoculate each test circle with a visible inoculum from an 18-72-hour, pure culture of yeast using an applicator stick (provided).
- Incubate at room temperature for five (5) minutes.
- Add one (1) drop of Bacticard® Candida Color Developer to the PRO test circle.
- Observe for a red color development within 30 seconds.
- Add one (1) drop of Bacticard® Candida MUGAL reagent to the MUGAL test surface.
- Observe the MUGAL test circle in a darkened room with a longwave ultraviolet light for bright blue fluorescence.
Interpretation
PRO test
Pos = red color development within 30 seconds
Neg = no color change
MUGAL test
Pos = bright blue fluorescence
Neg = no fluorescence
| Quality Control | PRO | MUGAL |
| Candida albicans | pos | pos |
| Crytococcus neogormans | neg | neg |
Notes
- Further testing is required for definitive identification.
- Testing cultures less than 18 hours old may result in false negatives.
- Incubating for longer than the recommended test reaction times may result in false positive results.
- Some non-candida species of yeast may fluoresce yellow on the MUGAL test.
Bacticard® Neisseria
Remel Bacticard® Neisseria
This test is only performed on oxidase-positive Gram-negative diplococci. Colonies should be 18-48 hours old from selective agar (Thayer-Martin, Martin-Lewis or New York City).
- Label Bacticard® Neisseria with the specimen identification.
- Inoculate only one isolate per test card.
- Add one (1) drop of Bacticard® Neisseria Rehydrating Fluid to each test circle.
- Inoculate each test circle with a visible inoculum of the test isolate using an applicator stick (provided in kit).
- Incubate at room temperature for two (2) minutes.
- Observe the IB test circle for a blue-green color indicating a positive IB test. If positive, do not proceed beyond this point. See below for interpretation.
- If the IB test is negative, continue incubation at room temp for 13 additional minutes (15 minutes total test time).
- Observe the BGAL test circle for a blue-green color indicating a positive test. If positive, do not proceed beyond this point. See below for interpretation.
- If the BGAL test is negative, proceed by adding one (1) drop of Bacticard® Neisseria Color Developer to the GLUT and PRO test circles.
- Observe GLUT and PRO test circles for development of a pink to red color at 30 seconds. See below for interpretation.
Interpretation
- IB test
- Pos = blue-green color development within 2 minutes
- Neg = no color change within 2 minutes
- BGAL test
- Pos = blue-green color development within 15 minutes
- Neg = no color change
- GLUT test
- Pos = pink to red color development within 30 seconds after the addition of Color Developer
- Neg = no color change
- PRO test
- Pos = pink to red color development within 30 seconds after the addition of Color Developer
- Neg = no color change
Quality Control
| Organism | IB | BGAL | GLUT | PRO |
|---|---|---|---|---|
| Moraxella catarrhalis | + | |||
| Neisseria lactamica | – | + | ||
| Neisseria gonorrhoeae | – | – | – | + |
| Neisseria meningitidis | – | – | + | – |
Notes
- Kingella spp. are Gram negative coccobacillary organisms which may be isolated on Thayer Martin medium. They can be differentiated from Neisseria gonorrhoeae by the superoxol test (30% H2O2).
- Neisseria cinerea can be PRO-positive but is rarely isolated from genital sites. It can be distinguished from Neisseria gonorrhoeae by its growth on nutrient agar or tryptic soy agar.
- Isolates recovered from primary isolation on nonselective media such as chocolate agar should only be tested after subculture to appropriate selective media. Most Neisseria spp. (N. sicca, N. mucosa, N. cinerea) possess prolyl aminopeptidase and may be confused with Neisseria gonorrhoeae if not tested properly. Further biochemical and serological testing may be required.
- Interpretation of the IB test past the 2-minute incubation time may result in false-positive reactions.
- PRO-negative strains of Neisseria gonorrhoeae and GLUT-negative strains of N. meningitidis have been reported. Additional testing such as carbohydrate acidification is required to definitively identify these isolates.
Bile Esculin Procedure
Oxoid™ Bile Esculin Slant
- Use a sterile stick or loop to inoculate 1-2 colonies from an 18-24-hour culture to the surface of the bile esculin slant. Use a fishtail motion to inoculate the surface of the slant from bottom to top.
- Incubate at 35-37°C in ambient air for 18-24 hours.
Pos = blackening of the slant
Neg = no blackening
Positive QC: Enterococcus sp.
Negative QC: Streptococcus pyogenes, Streptococcus agalactiae
BD™ Bile Solubility Test for Alpha-Hemolytic Streptococci
The bile solubility reagent consists of 10% aqueous sodium desoxycholate. The principle of the test is based on the fact that Streptococcus pneumoniae contains an autolytic enzyme, the action of which can be accelerated in the presence of a bile salt. All other streptococci are not bile soluble.
Direct Colony Procedure
- Place one (1) drop of reagent on a well-isolated alpha-hemolytic colony that is 18-24 hours old. Take note of exactly where the suspicious colony is on the plate (perhaps by marking the back of the plate).
- Incubate the plate at room temperature, agar surface up, for 30 minutes (leaving the plate slightly ajar may help speed up the drying process).
- Examine the appearance of the colony. The test is positive if the colony has disintegrated.
Positive = colony has disintegrated (Streptococcus pneumoniae)
Negative = no change in colony
Positive QC: Streptococcus pneumoniae
Negative QC: Alpha-hemolytic streptococci, not Streptococcus pneumoniae (e.g. viridians streptococci)
Blood Agar Staph Streak (BASS): Test for Satellitism
- Inoculate one colony of a suspected Haemophilus sp. to a sheep blood agar plate and streak for isolation.
- Place a single (or Y-shaped) streak line of a beta hemolysin-producing strain of
Staphylococcus aureus over the inoculated plate as demonstrated in the lab, preferably in the first and second quadrants.
- Incubate the plate in CO2 for 18-24 hours.
- After incubation, observe for the presence of small colonies close to the Staphylococcus aureus streak line that diminish in size farther away from it.
Quality control
Pos: Haemophilus influenzae
Neg: Bordetella parapertussis
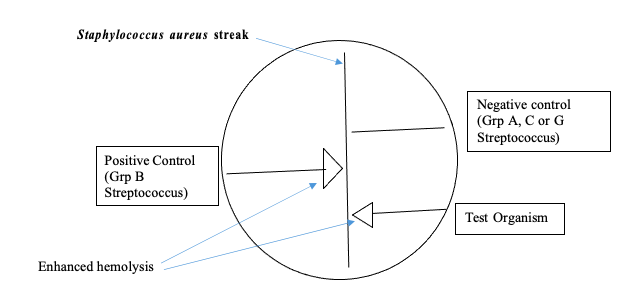
CAMP Test Procedure
CAMP Test – Waterfront
- Touch 1 colony of a beta-hemolytic Staphylococcus aureus with a sterile loop and streak it down the center of a blood agar plate (Note: Oxoid™ blood agar plates may not work well).
- Streak QC and unknown organisms perpendicular to the S. aureus streak stopping about 2-3 mm from it. Be careful to avoid touching the organism to the S. aureus streak line. Note: Several organisms can be tested on one plate as long as they are 3-4 mm apart from each other.
- Incubate the plate in ambient air at 35-37°C overnight.
- Observe for an arrow-shaped area of enhanced hemolysis at the junction of the unknown organism and the S. aureus.
Pos = arrowhead-shaped zone of enhanced hemolysis at junction between organism and aureus
Neg = no enhancement of hemolysis
Positive QC: Streptococcus agalactiae
Negative QC: Streptococcus pyogenes
Catalase Test Procedure
Catalase – Waterfront
- Use a sterile stick or loop (not nickel chromium) to transfer 1 or 2 isolated colonies to the surface of a clean microscope slide. (Note: be careful not to “dig” the agar of a blood agar plate. This can cause a false positive reaction due to the pseudo-peroxidase nature of blood).
- Place a drop of 3% hydrogen peroxide on the organism(s).
- Observe for rapid production of bubbles (effervescence).
Pos = rapid production of bubbles
Neg = weak or no production of bubbles
Note
Positive QC: Staphylococcus sp.
Negative QC: Streptococcus sp.
- Alternatively, a drop of hydrogen peroxide can be placed on a slide first and then a colony can be added to it.
- Enterococcus sp. may give a weak positive (pseudo catalase) reaction when grown on blood agar. A truly catalase-positive organism will produce a rapid effervescence.
Catarrhalis Test Disk
The enzyme, butyrate esterase, releases indoxyl from indoxyl butyrate and spontaneously forms indigo in the presence of oxygen.
Remel™ Catarrhalis Test Disk
- Gram-stain the isolate prior to performing the test. Test only oxidase-positive, Gram negative diplococci.
- Using forceps, place the disk on a clean glass microscope slide. Do not rehydrate the disk.
- Rub several colonies (a visible inoculum) across the disk using a wooden applicator stick.
- Observe for a blue-green color development within 2 minutes.
Results
- Pos = blue-green color within 2 minutes
- Neg = no color development within 2 minutes
Quality Control
| Moraxella catarrhalis | pos |
| Neisseria lactamica | neg |
Note: this test should only be used for the identification of Moraxella catarrhalis from clinical samples.
Chlamydospore Production
Corn meal agar with added Tween 80 is used as a substrate for chlamydospore production. Chlamydospores are asexual fungal spores that are derived from a hyphal cell and can function as a resting spore.
- Use a straight wire to pick one colony of yeast from a Sabouraud Dextrose agar plate and make a deep cut in the corn meal agar (i.e. a horizontal furrow). Repeat for each organism being tested.
- Place a flamed sterile cover slip over the line of inoculum.
- Incubate for 24-48 hours at 22°C.
- Examine the streaks microscopically, through the cover slip, using a low power objective.
Interpretation
Candida albicans produces mycelium-bearing ball-like clusters of budding cells and thick-walled round chlamydospores
Quality Control
Candida albicans: growth; production of budding cells with chlamydospores
Trichophyton mentagrophytes: growth
Citrate Utilization Test Procedure OXOID® Simmons Citrate Agar
- Touch an isolated colony with a sterile loop or stick and inoculate the surface of the slant using a fishtail motion.
- Incubate the tube with a loosened lid in ambient air at 35-37°C for 24-48 hours.
- Observe for color change at 24 hours.
- If negative, re-incubate for an additional 24 hours.
Pos = Bright blue color
Neg = Absence of growth with no change in the original green color
Positive QC: Klebsiella pneumoniae
Negative QC: Escherichia coli
Notes
- The reaction requires oxygen so inoculate the surface of the agar rather than stabbing it. Failure to incubate with a loose lid may also result in a false negative test.
- When setting up a battery of biochemicals from the same culture, inoculate this medium lightly before inoculating other media to avoid false positive reactions caused by carryover of glucose or other nutrients from other media.
Coagulase Test Procedures
Production of coagulase is indicative of a Staphylococcus aureus strain. The enzyme acts within host tissues to convert fibrinogen to fibrin. It is theorized that the fibrin meshwork that is formed by this conversion surrounds the bacterial cells or infected tissues, protecting the organism from nonspecific host resistance mechanisms such as phagocytosis and the anti-staphylococcal activity of normal serum.
Slide Coagulase – Waterfront
- Use a grease pencil (china marker) to draw two large ovals or rectangles on a labeled glass microscope slide. Label one “S” and the other “S&P”
- Place one drop of sterile saline in each circle on the slide. Use a sterile stick or loop to remove several (about 10 or more) isolated colonies from a plate and then emulsify them in both saline drops. Note: a smooth, fairly heavy suspension is required. If the suspension clumps, do not proceed with the next step.
- Add one drop of rabbit plasma (non-citrated) to the circle labeled “S&P”. Mix briefly with a fresh stick or sterilized loop.
- Mix or rock the slide gently for 5-10 seconds and observe for signs of macroscopic clumping
Pos = macroscopic clumping in the rabbit plasma in ≤ 10 seconds with no clumping in the saline
Neg = No clumping in the saline drop or the plasma
Notes
- A negative slide coagulase test must always be confirmed with a tube coagulase test.
- Any clumping in the saline indicates autoagglutination. This organism cannot be reliably tested with the slide coagulase method.
- The use of sodium citrate as an anticoagulant in the collection of the rabbit plasma will result in false positive reactions. EDTA is an acceptable anticoagulant.
- S. lugdenensis and S. schleiferi may give positive slide coagulase reactions so positive results should always be confirmed by a tube coagulase test
Positive QC – Staphylococcus aureus
Negative QC – Staphylococcus epidermidis
Staphaurex® Test
Staphaurex® is a commercial product that is used to check staphylococci for the presence of bound coagulase and Protein A. This makes it more sensitive than the slide coagulase test which checks for bound coagulase only.
Staphaurex® is a latex agglutination test that has replaced the slide coagulase test in many laboratories. The reagent contains polystyrene latex particles which have been coated with fibrinogen and immunoglobulin G (IgG). Bound coagulase (clumping factor) reacts with the fibrinogen to cause clumping. Protein A (found in 95% of Staphylococcus aureus strains) has an affinity for the Fc portion of IgG, which also results in strong agglutination of the latex particles. This test gives about 99% sensitivity and specificity for the presence of Staphylococcus aureus.
Staphaurex® Test Procedure
- Remove kit from the fridge and allow reagent to come to room temperature before use.
- Shake the reagent vigorously and examine for aggregation before use. If there are signs of lumpiness, do not use the reagent.
- Holding the reagent bottle vertically, place one (1) drop of latex reagent in the circle on the reaction card.
- Use a sterile stick to pick up about six well-isolated colonies from a fresh blood agar, nutrient agar or CNA plate which have stained as Gram positive cocci (staphylococcal-like).
- Emulsify the colonies in the drop of latex reagent on the reaction card. Spread out the reagent over about half the area of the circle. Discard the stick in biohazard garbage. Make note if the colonies did not emulsify smoothly.
- Rock the card gently for up to 20 seconds and examine for agglutination (do not use a magnifying lens to visualize).
Positive = clearly visible clumping of the latex particles with clearing of the milky background. Negative = latex does not agglutinate, and the milky appearance does not change
Notes
- Results should be confirmed with a tube coagulase test.
- Increased granularity may be seen if you rotate the card more than 20 seconds.
- Rough or stringy reactions appear as white specks or stringy aggregates and should be interpreted as follows:
- Negative, if accompanied by a milky background
- Positive if accompanied by a clear background. Hint: I often record these as “weak positive” or “positive?” and perform a tube coagulase test to confirm.
- Specimens grown on high-salt media, e.g. MSA, often give rough or stringy emulsions with weak positive reactions
- Staphylococcus intermedius and Staphylococcus hyicus may give positive results but are seldom clinically significant. If necessary, they can be distinguished from S. aureus with PYR and VP tests. Staphylococcus saprophyticus may also give a weak positive reaction but it will be negative in the tube coagulase test.
- Other organisms such as E. coli and Candida albicans may non-specifically agglutinate latex particles. Ensure that your colonies have a staphylococcal morphology before performing this test on them.
Positive QC: Staphylococcus aureus
Negative QC: Staphylococcus epidermidis
Decarboxylase Test Procedure
OXOID® Moeller Decarboxylase (e.g. Ornithine, Lysine) Or Dihydrolase (e.g. Arginine) Procedure
- Inoculate each broth medium (e.g. ornithine test, arginine test, lysine test) with organisms from an 18-24-hour culture using a sterile loop, stick or straight wire.
- A basal control tube (labelled as “decarboxylase base”) should also be inoculated at the same time.
- Overlay all tubes, including the base (control), with 1-2 mL of sterile mineral oil.
- Incubate the tubes in ambient air at 35-37oC for 18-24 hours.
- Observe for colour change and re-incubate up to 4 days if test is negative, checking each day for any color change.
Pos = purple (or purple yellow) “test” tube and yellow base (with apparent turbidity)
Neg = yellow “test” and yellow base for glucose fermenters; little or no color change in comparison to an uninoculated tube for non-fermenters (yellow brown)
| QC | Ornithine | Lysine | Arginine |
|---|---|---|---|
| Klebsiella pneumoniae | neg | pos | neg |
| Enterobacter cloacae | pos | neg | pos |
Notes
- These tests rely on the ability of the organism to ferment glucose to initiate decarboxylation. Any organism that does not have the ability to utilize glucose in the absence of oxygen may not give reliable test results. The control (or base) tube containing no amino acid should turn to yellow and remain that color if the organism being tested ferments glucose after 18-24 hours of incubation. A purple-coloured control/base tube invalidates all the amino acid decarboxylase tests and no interpretation should be made in this case.
- Results of the decarboxylase tests cannot be interpreted until the tubes have been incubated a minimum of 18 hours.
- A positive decarboxylase test may be difficult to interpret due to the presence of an indistinct yellow-purple color. If this occurs, always compare with an uninoculated tube. Any trace of purple color denotes a positive test if the tube has been incubated at least 18 hours.
- Some organisms may be slow in decarboxylation and may require increased incubation of up to 4 days.
- The addition of mineral oil provides an anaerobic environment (required for decarboxylation) and prevents oxidation which would cause a pH rise and hence, a false positive reaction.
DNase Test Procedure
OXOID® DNase Agar with Methyl Green
- Touch an isolated colony from an 18-24-hour culture with a sterile loop and inoculate to the surface of the agar plate with a wide streak (width of loop) that is at least 2.5 cm long or in a circular spot about 5-6 mm in diameter.
- Incubate the plate in ambient air at 35-37°C for 18-24 hours.
- Observe for a zone of yellow clearing in the agar surrounding the growth.
Pos = yellow clearing around the colonies
Neg = agar surrounding colonies remains green
Positive QC: Serratia marcescens
Negative QC: Escherichia coli
Notes
- Staphylococcus aureus is DNase positive and can be used as an alternative positive control.
- Moraxella catarrhalis is also DNase positive but it may grow poorly on media containing indicator dyes, e.g. methyl green, toluidine blue.
- Klebsiella pneumoniae is DNase negative and can be used as an alternative negative control.
Germ Tube Procedure
Remel Germ Tube Solution
Formation of germ tubes is associated with increased synthesis of protein and ribonucleic acid. Remel germ tube solution contains tryptic soy broth and fetal bovine serum, which provide essential nutrients for protein synthesis.
- Make a dilute suspension by touching one colony of an unknown yeast and transferring it to a tube of germ tube solution. DO NOT inoculate the solution heavily. Excessive inoculum causes a significant decrease in the percentage of cells forming germ tubes.
- Incubate aerobically at 35-37°C for 2-4 hours.
- Examine microscopically under 40X magnification for the presence of germ tubes.
Interpretation
Pos = a short hyphal (filamentous) extension arising laterally from a yeast cell, with no constriction at the point of origin
Neg = no hyphal (filamentous) extension arising from a yeast cell or a short hyphal extension constricted at the point of origin
Quality Control
| Candida albicans | pos |
| Candida glabrata | neg |
Notes
- Candida dubliniensis also produces germ tubes and chlamydospores. It can be differentiated from Candida albicans by testing for growth at elevated temperatures and colony morphology on differential media.
- Candida triopicalis may produce early pseudohyphae that can be confused with germ tubes. Its hyphal extensions, however, are constricted at the point of origin.
- Further testing is required for definitive identification.
Gram Stain Procedure
Smear preparation
- Use a pencil to label the frosted end of a microscope slide. Include the patient’s identification and your initial. Draw a circle or oval in the middle or near the far end of the slide with a grease pencil.
- Fluid sample: Add a drop of fluid to the circle with a sterile pipette or loop and allow to air-dry.
- Colony: Place a drop of sterile saline in the circle. Use a sterile straight wire, loop or stick to touch one colony on a culture plate and then gently emulsify the colony in the saline on the slide. Set the slide aside to air-dry.
- Methanol fix smears for one minute, then let the slide air dry.
Staining
- Place slides on a staining rack in the sink
- Gently flood the slide with crystal violet and wait for 1 minute. Wash well with tap water
- Apply Gram’s iodine for 1 minute. Wash well with tap water.
- Decolorize carefully with acetone-alcohol until no more purple color washes out. Immediately rinse well with water.
- Counterstain with 1% safranin for 1 minute. Wash well with water.
- Blot and allow to air-dry
Reading Gram-stained smears
- Examine under 10X first. Focus on any possible bacterial or fungal structures.
- Add a drop of immersion oil and re-focus on 10X. Move from the 10X objective directly to the 100X objective (oil immersion lens) and re-focus. Do not drag the 40X objective though the immersion oil on the slide!
- Always record your results when you read them. Do not rely on your memory!
- Slides can be discarded in biohazard garbage or stored in a cardboard slide tray after gently blotting off most of the oil.
- Correlate the Gram stain results to your colony morphologies and the growth on the different types of media.
Hippurate Procedure
Remel® Hippurate Disk Test
- Add 0.1 mL (2 drops) of demineralized water to a small test tube.
- Emulsify 1-3 colonies of a fresh isolate in the water.
- Using forceps drop a hippurate disk into the suspension.
- Incubate aerobically at 35-37°C. (Best practice would be for 2 hours, but we will try overnight incubation)
- Dispense 2 drops of ninhydrin reagent into the test tube.
- Re-incubate aerobically at 35-37°C for 30 minutes.
- Observe for a blue to purple color development
Pos = Deep blue or deep purple color Neg = Colorless or slightly yellow pink
Positive QC: Streptococcus agalactiae
Negative QC: Streptococcus pyogenes
Indole (Spot) Test Procedure
BD™ DMACA Indole Reagent Droppers:
- Hold the reagent dropper upright and point the tip away from yourself. Grasp the middle of the dropper with your thumb and forefinger and squeeze gently to crush the ampule inside the dropper. Caution: break ampule close to its center one time only. Tap bottom of dropper on tabletop a few times.
- Place a small piece of filter paper on a glass microscope slide or in a Petri dish.
- Invert the dropper and squeeze the center to add a drop or two of reagent to the filter paper. Caution: do not over-saturate the filter paper (it is best if you can see a dry area on the outer edge of the filter paper). Over-saturation may result in false negative reactions.
- Use a sterile loop or wooden stick to select a well-isolated isolated colony from an 18-24- hour culture grown on a blood agar plate that contains tryptophan, e.g. trypticase soy agar with 5% sheep blood.
- Rub the colony onto the reagent-saturated portion of the filter paper.
- Observe for color change within 2 minutes.
Pos = blue to blue green
Neg = no color to pink
Positive QC: Escherichia coli
Negative QC: Klebsiella pneumoniae, Pseudomonas aeruginosa
Notes
- Colonies to be tested must be isolated on media containing sufficient tryptophan and no glucose. Trypticase soy agar is a good example.
- Allow at least 2 minutes for the absence of a reaction before calling the test negative.
- Colonies should not be tested from media containing dyes. Dye carryover can result in erroneous results.
- Colonies from mixed cultures should not be tested. Indole-positive colonies may cause nearby (within 5 mm) indole-negative colonies to appear weakly positive.
- False negatives may occur with some organisms due to rapid breakdown of indole, e.g. Clostridium
- Each dropper is good for one day’s use after the ampule has been broken.
- DMACA indole reagent is very acidic. Avoid contact with skin, eyes and mucous membranes. Rinse thoroughly with water if spilled.
- Do not use the reagent if the color is brown (indicates deterioration)
Indoxyl Acetate Disk Test
Bacterial esterase, releases indoxyl from indoxyl acetate and spontaneously forms indigo in the presence of oxygen.
Remel™ Indoxyl Acetate Disk Test Procedure
- Using forceps, place the disk on a clean glass microscope slide.
- Rehydrate the disk with one drop of demineralized water; do not oversaturate.
- Use an applicator stick to apply several 18-72-hour old colonies of the test isolate to the disk.
- Incubate the disk at room temperature for 20 minutes.
- Examine for a blue or blue-green color development.
Results
Pos = blue or blue-green color
Neg = no color development
Quality Control
| Campylobacter jejuni ATCC 33291 | pos |
| Campylobacter fetus subsp. fetus ATCC 27374 | neg |
Kirby-Bauer Disc Diffusion Susceptibility Method
- Bring all plates, required antibiotics and saline to room temperature.
- Label a Mueller Hinton agar plate and blood agar plate with the date and time
- Touch four to five well-isolated colonies of the same colony morphology (18-24-hour colonies from a non-selective medium) with a sterile swab.
- Aseptically transfer the swab containing the organism to a tube of sterile saline and rotate the swab in the saline until the turbidity is visually equivalent to a 0.5 McFarland standard.
- Mix well by gently and slowly tapping the bottom of the tube several times with your fingers.
- If your suspension is more turbid than the 0.5 McFarland turbidity standard, carefully dilute the tube by adding more sterile saline. DO NOT OVERFILL THE TUBE! Mix and compare with the turbidity standard.
- If you cannot adjust the turbidity in this way, repeat steps 1-5 using a fresh tube of sterile saline.
- Within 15 minutes of inoculum preparation, dip a fresh sterile cotton swab into the well- mixed suspension and express the excess liquid by rotating the swab firmly against the inside of the tube above the fluid level.
- Inoculate the Mueller-Hinton agar plate by streaking the swab over the entire agar surface. Rotate the plate 60o and streak the entire surface again with the same swab. Repeat this step one more time so that a confluent lawn of growth will be achieved over the entire surface. A final sweep of the agar surface with the swab against the rim will ensure adequate coverage.
- Use the same swab to inoculate a blood agar plate (labeled with the specimen identification and the words “purity plate”) and then discard the swab into a biological waste container. Use a sterile loop to streak the purity plate for isolated colonies.
- Allow the Mueller Hinton plate to dry (inverted so that no condensation drops onto the agar) for about three minutes.
- Remove the discs you need from the antimicrobial cartridges using sterilized forceps or a dispenser. Place the discs onto the lid of the Mueller Hinton plate (only if no condensation has collected there) or into a sterile Petri dish. Do not allow the discs to come into contact with moisture or with each another.
- Aseptically place each disc on the inoculated Mueller Hinton agar plate with the discs spaced equidistant from each other and no closer than 24 mm from each other (center to center). They should also be at least 15 mm from the edge of the plate. You can place a maximum of six (6) discs on a 100 mm plate or twelve (12) discs on a 150 mm plate. It is very important that discs be placed onto the plate surface within 15 minutes of organism inoculation. Do not move the discs once they have touched the plate surface.
- With forceps or a sterile stick, tap the top of each disk to ensure adherence of the disks to the agar surface.
- Within 15 minutes of disc placement, invert and incubate the Mueller-Hinton agar plate at 35° C in an ambient air incubator for16-24 hours (Duration of incubation depends on CLSI protocol for that organism. For example, the required incubation for Enterobacteriaceaesp. except for oxacillin and vancomycin MIC’s for all staphylococci and cefoxitin discs on coagulase negative staphylococci which must be read at 24 hours).
After incubation (Day 2)
- After incubation, evaluate the purity plate to ensure that the organism suspension used for the test was pure. To do this, ensure that the colony morphology on the plate matches how you described the organism on the previous day. Also check for more than one colony morphology. If it looks pure, record the purity plate result as “pure” or “OK” or describe it using the same terms that were used on the previous day. If it is not pure, the zone sizes cannot be read.
- Check the Mueller Hinton plate for confluent growth. If individual colonies are apparent, the inoculum was too light, and the test should be repeated.
- Read the results of the Mueller Hinton plate by measuring the diameter of the zones of complete inhibition around the disc through the bottom of the plate. Remember to use reflected light and a dark background and to measure through the diameter of the disc. Record zone sizes to the nearest mm.
- If there is no zone of inhibition, record the zone size as ≤ 6 mm.
- Refer to the appropriate CLSI Zone Diameter Interpretive Chart to determine if each organism is “S” (susceptible), “SDD” (susceptible-dose dependent), “I” (Intermediate) or “R” (resistant) to each antibiotic. Record the interpretation next to each antibiotic zone size on your worksheet.
Notes
- If the purity plate or the Mueller Hinton plate appears to have more than one colony form, each colony form should be sub-cultured and identification tests should be repeated on each one.
- If scattered colonies occur inside the zone of inhibition (other than with sulphonamides and trimethoprim), the organism should be checked for purity. If these colonies end up being the same identity as the isolate being tested, the organism should be interpreted as resistant to that antibiotic. (This happens when resistant mutants are produced in the presence of the antibiotic.)
- When interpreting the results for Proteus spp. which swarm, disregard any swarming which occurs between the margin of heavy growth and the disc.
- When reading results for sulphonamide and/or trimethoprim discs, a haze of growth within the zone of inhibition can be ignored since the organism may grow through several generations before inhibition occurs. Measure the zone from the margin of heavy growth.
- When reading sensitivity results on penicillin and ampicillin for staphylococci, observe the appearance of the colonies at the edge of the zone of inhibition. If there is a haze of growth within the zone of inhibition, check for MRSA.
Lecithinase and Lipase Production
Egg Yolk Agar is used for the detection of Lecithinase and lipase by bacteria.
- Inoculate the test organism on the surface of an Egg Yolk agar plate and incubate aerobically (for aerobes) or anaerobically (for anaerobes) for 24-48 hours.
- Examine the plate for production of a white opaque zone surrounding a colony or an iridescent sheen on the surface of the agar when the plate is held at an angle under a light source.
Interpretation
White opaque zone extending from edge of colony = lecithinase positive
Iridescent sheen on agar surface = lipase positive
Quality Control
Bacteroides fragilis: growth; lecithinase neg
Clostridium perfringens: growth; lecithinase pos
Clostridium sporogenes: growth; lipase pos, white zone
Mannitol Salt Agar Procedure
Mannitol salt agar
This medium is selective for salt-tolerant organisms such as staphylococci. Differentiation among the staphylococci is predicated on their ability to ferment mannitol. Following incubation mannitol-fermenting organisms, typically S. aureus strains, exhibit a yellow halo surrounding their growth while non-fermenting strains produce colonies with reddish purple or reddish pink zones.
Oxoid™ Mannitol Salt Agar
- For clinical specimens, inoculate directly to the media surface and streak for isolated colonies.
- For colonies from culture media, touch an isolated colony with a sterile loop and inoculate the surface of the agar. Streak for isolated colonies.
- Incubate the plate in ambient air at 35-37°C for 24 hours.
- If negative, re-incubate the plate for an additional 24 hours.
Pos = growth of smooth, raised colonies with a yellow color change in the medium Neg = absence of growth or inhibited growth or growth of smooth, raised colonies with a red-pink zone around the colonies.
Positive QC: Staphylococcus aureus –
Negative QC: Staphylococcus epidermidis – growth with pink zones at 48 hours
Proteus mirabilis – inhibition of growth
Notes
- While this medium is selective for pathogenic staphylococci, a coagulase test must be performed for confirmation of the organism identification.
- Some strains of streptococci/enterococci and staphylococci will grow on this medium and ferment mannitol.
Motility-Indole-Lysine Test Procedure
OXOID® Motility-Indole-Lysine (MIL) Medium
- Touch an isolated colony or several identical colonies from an 18-24-hour culture with a sterile straight wire and stab the center of the MIL medium to about ½ its length. Try to keep your stab line as narrow as possible.
- Incubate the tube with a loosened lid in ambient air at 35-37°C for 18-24 hours.
- After incubation, first observe the tube for motility and lysine decarboxylation.
- Add 3-4 drops of Kovac’s indole reagent and observe for indole production.
Motility Interpretation
Pos = diffuse turbidity or visible growth outward from the stab line
Neg = no turbidity or visible growth only along the stab line.
Lysine interpretation
Pos = purple throughout tube
Neg = yellow
Indole Interpretation
Pos = pink to red color on the top of the medium after the addition of Kovac’s reagent.
Neg = no color change/yellow
Positive QC: Escherichia coli
Negative QC: Klebsiella pneumoniae
Notes
- Lysine and motility tests must be interpreted and recorded before adding Kovac’s reagent. (The reagent will change the colour and cause clouding of the medium)
- Motility tests may show a false negative reaction if organisms are weakly motile or if the flagella are damaged due to heating, shaking or other trauma. A wet-prep motility test from an inoculated tryptone broth may be performed to confirm motility results.
- Oxidation may cause a purple color near the top of the tube. If the rest of the tube is yellow, the lysine test should be considered negative.
- Failure to incubate with caps loose may produce erroneous results.
- If DMACA is used as the indicator instead of Kovac’s reagent, a positive reaction will be blue rather than pink-to-red.
Nitrate Reduction Test Procedure
OXOID™ Nitrate Broth with Durham Tube
- Touch an isolated colony with a sterile loop or stick and inoculate the nitrate broth.
- Incubate the tube in ambient air at 35-37°C for 18-24 hours.
- To the incubated tube add 5 drops each of alpha-naphthylamine (Nitrate A) and sulfanilic acid (Nitrate B). Mix well.
- Observe for the production of a red color within 1-2 minutes.
- If no color is observed, proceed with the next two steps.
- Examine the inverted Durham tube for the presence of bubbles (indicates gas production).
- Dip a sterile stick into zinc dust and transfer only the amount that sticks to the stick to the tube. Observe for the production of a red color within 5-10 minutes. (Note: the stick can be broken off and left in the aliquot tube to help you remember what step you are at in the process).
Interpretation
Deep red color after addition of Nitrate A and Nitrate B = positive for reduction of nitrates to nitrites.
No color change after the addition of Nitrate A and Nitrate B and a deep red colour after the addition of zinc dust = negative for reduction of nitrates.
No color change after addition of Nitrate A and Nitrate B, gas in the Durham tube, and no color change after addition of zinc dust = pos for reduction of nitrates to nitrites and then to nitrogen gas.
Quality Control
Positive QC: Escherichia coli: nitrate reduced, gas negative
Pseudomonas aeruginosa: nitrate reduced; gas positive
Negative QC: Acinetobacter spp.: Nitrate not reduced, gas negative
Notes
- The diazonium compound produced by the reaction of nitrite with reagents A and B is unstable and the color may fade. Color reactions should therefore be read shortly after adding reagents.
- Tubes containing a Durham tube should be inverted before inoculation to dislodge any trapped air bubbles in the Durham tube. Caution: do this only with tubes that have a screw cap lid and are tightly closed!
- A slight pink color may be produced after the addition of nitrate reagents. This is not a positive test. A positive reaction is indicated by a deep red color.
- Sometimes the reaction is incomplete. When this occurs, there may be gas in the Durham tube and a red color is produced after the addition of nitrate reagents. A longer incubation time may show more complete nitrate reduction.
- The test cannot be re-incubated after the addition of reagents so removal of an aliquot for testing is recommended. The original broth can be re-incubated up to 5 days.
- Some strains may completely reduce nitrate but will not produce gas.
Nitrate Disk (Anaerobic)
Remel Nitrtae Disk (Anaerobic)
- Allow disks to warm to room temperature.
- Select one well-isolated colony from a nonselective anaerobic blood agar plate and streak the first quadrant of the plate back and forth several times to ensure an even lawn of growth. Streak the other quadrants for isolation.
- Place the nitrate disk on the heavily inoculated part of the plate using forceps. Subsequent discs for identification can be placed on the plate.
- Aerotolerance testing should be performed at the same time if not already done.
- Incubate the plate anaerobically at 35-37oC for 24-48 hours (or until good growth of the organism is evident).
- Remove disk from surface of plate and place in a clean dry petri dish.
- Add one drop each of Anaerobic Nitrate Reagents A and B to the disc. If no color develops within 3-5 minutes, add a small amount of zinc dust and wait 5 minutes. Observe for color change.
Interpretation
Pos = red color development after addition of Nitrate A and B; no color change after the addition of zinc dust.
Neg = No color change after the addition of Anaerobic Nitrate A and B; red color development after addition of zinc dust
| Quality Control | ||
|---|---|---|
| Bacteroides fragilis group: | neg | |
| Bacteroides ureolyticus-like | group: | pos |
| Fusobacterium spp. | neg | |
| Veillonella spp. | pos | |
| Actinomyces israelii | pos | |
| Propionibacterium acnes | pos | |
Notes
- A rapidly growing organism may turn the disc a tan color. When the reagents are added, no color or a very subtle color change may occur. Perform a tube test instead.
- If after 48 hours of incubation there is little or no growth, re-incubate before adding reagents.
Nitrocefin Disk Test Procedure
Remel® Nitrocefin Disk Test procedure
This procedure is used to detect the presence of beta-lactamase. It is typically performed on colonies of Staphylococcus spp., Haemophilus influenzae, Neisseria gonorrhoeae, Moraxella catarrhalis, Enterococcus sp. and anaerobic bacteria. As beta-lactamase enzymes become more common in these organisms, the test’s usefulness will lessen, and full antimicrobial susceptibility testing will eventually become routine.
In the test a paper disc is impregnated with a chromogenic cephalosporin called nitrocefin. The color of the nitrocefin changes from yellow to pink when the amide bond in its beta-lactam ring is hydrolyzed by a beta-lactamase,
Procedure
- Dispense a nitrocefin disc onto a clean microscope slide or an empty petri dish lid.
- Moisten the disc with one loopful of sterile demineralized water. Do not flood the disc.
- Use a sterile stick, toothpick or loop to smear 5-6 well-isolated colonies onto the disc surface.
- Observe the disc for a color change at the appropriate time as indicated below: Organism Reaction Time
Staphylococcus aureus 1 hour Haemophilus influenzae 5 minutes Neisseria gonorrhoeae 5 minutes Enterococcus faecalis 5 minutes Anaerobic bacteria 30 minutes
Note: If your organism is different than those listed above, use the reaction time for the genus that is most similar to your organism. For example, use the reaction time for Staphylococcus aureus if you are testing a coagulase-negative Staphylococcus.
Results
Positive = Red or pink color on area of disc where colonies were applied
Negative = Pale yellow or no color change
Novobiocin Susceptibility Test Procedure
Used to distinguish Staphylococcus saprophyticus from the rest of the clinically significant coagulase negative staphylococci:
- Bring a Mueller Hinton plate and novobiocin discs to room temperature.
- Touch four to five well-isolated colonies of the same colony morphology (18-24-hour colonies from a non-selective medium) with a sterile swab.
- Aseptically transfer the swab containing the organism to a tube of sterile saline and rotate the swab in the saline until the turbidity is visually equivalent to a 0.5 McFarland standard.
- Mix well by gently and slowly tapping the tube several times with your fingers.
- If your suspension is more turbid than the 0.5 McFarland standard, carefully dilute the tube by adding more sterile saline. DO NOT OVERFILL THE TUBE! Mix and compare with the turbidity standard.
- If you cannot adjust the turbidity in this way, repeat steps 1-3 using a fresh tube of sterile saline.
- Within 15 minutes of inoculum preparation, dip a fresh sterile cotton swab into the well- mixed suspension, rotate it several times and express the excess liquid by rotating the swab firmly against the inside of the tube above the fluid level.
- Inoculate the Mueller-Hinton agar plate by streaking the swab over the entire agar surface. Rotate the plate 60o and streak the entire surface again with the same swab. Repeat this step one more time so that a confluent lawn of growth will be achieved over the entire surface. A final sweep of the agar surface with the swab against the rim will ensure adequate coverage.
- Use the same swab to inoculate a blood agar plate (labeled with the specimen identification and the words “purity plate”) and then discard the swab into a biological waste container. Use a sterile loop to streak the purity plate for isolated colonies.
- Allow the Mueller Hinton plate to dry (inverted so that no condensation drops onto the agar) for about three minutes.
- Remove a 5ug novobiocin disc from the antimicrobial cartridge using heated forceps or a dispenser. Place the disc onto the lid of the Mueller Hinton plate (only if no condensation has collected there) or into a sterile Petri dish. Do not allow the disc to come into contact with moisture or another type of antibiotic.
- Aseptically place the disc on the inoculated Mueller Hinton agar plate with the disc spaced at least 15 mm from the edge of the plate. Do not move the disc once it has touched the agar surface. It is very important that discs be placed onto the plate surface within 15 minutes of organism inoculation.
- With forceps or a sterile stick, tap the top of the disk to ensure adherence to the agar surface.
- Within 15 minutes of disc placement, invert and incubate the Mueller-Hinton agar plate at 35 ± 2° C in an ambient air incubator for 18-24 hours.
- After incubation, evaluate the purity plate to ensure that the organism suspension used for the test was pure. To do this, ensure that the colony morphology on the plate matches how you described the organism on the previous day. Also check for more than one colony morphology. If it looks pure, record the purity plate result as “pure” or “OK” or describe it using the same terms that were used on the previous day. If it is not pure, the novobiocin test cannot be read.
- Check the Mueller Hinton plate for confluent growth. If individual colonies are apparent, the inoculum was too light and the test should be repeated. Read the result of the Mueller Hinton plate by measuring the diameter of the zone of complete inhibition around the disc through the bottom of the plate. Remember to use reflected light and a dark background and to measure through the diameter of the disc.
- If there is no zone of inhibition, record the zone size as ≤ 6mm.
Interpretation
Susceptible = zone size >16 mm
Resistant = zone sizes ≤ 16 mm
Quality Control
Resistant to novobiocin: Staphylococcus saprophyticus
Susceptible to novobiocin: Staphylococcus epidermidis
Notes
- If the purity plate or the Mueller Hinton plate appears to have more than one colony form, each colony form should be sub-cultured and identification tests should be repeated on each one.
ONPG Test Procedure
Oxoid® DD0013™ Discs for Rapid Detection of Beta-galactosidase
- Place one disc into a labelled 12×75 mm test tube.
- Add 0.1 mL sterile saline (0.85%).
- Use a sterile stick or loop to pick one colony of unknown organism and emulsify it in the tube containing the disc and saline.
- Incubate at 35°C.
- Examine at hourly intervals for up to 6 hours. If negative at 6 hours, re-incubate for up to 24 hours to check for late lactose fermenters.
Pos = yellow Positive QC: Escherichia coli Neg = no colour Negative QC: Morganella morganii
BD® BBL™ Taxo™ Discs for Detecting Lactose Fermenters
- Place 0.5 mL of sterile trypticase soy broth in a labelled 12×75 mm test tube.
- Inoculate the broth with a heavy inoculum of organism taken from a TSI tube or a plate.
- Aseptically add an ONPG disc to the broth.
- Cover the tube and incubate in ambient air at 35-37°C for 4-6 hours.
- Observe for yellow color. Re-incubate up to 24 hours if negative.
Pos = yellow Positive QC: Escherichia coli Neg = no colour Negative QC: Morganella morganii
Notes
- A heavy inoculum is required in order to obtain a high concentration of enzyme and speed the reaction.
- The test cannot be used with yellow-pigmented bacteria unless the suspension is first centrifuged before interpreting results.
Optochin Susceptibility Test Procedures
BD BBL™ Taxo™ P Procedure (Non-standardized)
- Use a sterile loop to heavily inoculate 2-3 isolated alpha hemolytic colonies from an 18- 24 hour culture to a 5% sheep blood agar plate.
- Using heated forceps, place an optochin disk on the plate. Gently tap the disk to ensure adequate contact with the surface of the agar.
- Incubate at 35-37°C in 5% CO2 for 18-24 hours.
- After incubation, measure the zone of inhibition to the nearest millimetre.
Alternative Procedure (Standardized)
- Prepare a saline suspension of the suspect organism that is equivalent to a 0.5 McFarland turbidity standard.
- Use a swab to inoculate the suspension in 3 planes to half of a 5% sheep blood agar plate.
- Streak for isolated colonies on the other half of the plate.
- Using heated forceps, place an optochin disk in the heavily inoculated area of the plate. Gently tap the disk to ensure adequate contact with the surface of the agar.
- Incubate at 35-37°C in 5% CO2 for 18-24 hours.
- After incubation, measure the zone of inhibition.
Note
This procedure works well if you also want to add an oxacillin disk to screen for penicillin resistance in pneumococci
Pos: zone size ≥ 14 mm
Neg = no zone of inhibition
Equivocal: zone size < 14 mm. ID of Streptococcus pneumoniae is questionable. A bile solubility test must be performed to confirm the ID.
Positive QC: Streptococcus pneumoniae
Negative QC: Streptococcus mitis (or other alpha-hemolytic streptococcus)
BD™ Oxidase Test Procedure
BD™ Oxidase Reagent Dropper Method
- Hold the reagent dropper upright and point the tip away from yourself. Grasp the middle of the dropper with your thumb and forefinger and squeeze gently to crush the ampule inside the dropper. Caution: break ampule close to its center one time only. Tap bottom of dropper on tabletop a few times.
- Place a small piece of filter paper on a glass microscope slide or in a Petri dish.
- Invert the dropper and squeeze the center to add a drop or two of reagent to the filter paper. Caution: do not over-saturate the filter paper (it is best if you can see a dry area on the outer edge of the filter paper). Over-saturation may result in false negative reactions.
- Use a sterile platinum loop or wooden stick to select one or more isolated colonies from an 18-24-hour culture grown on non-selective, non-differential media. (Steel, nichrome or iron-containing loops may cause false positive reactions).
- Rub the colonies onto the reagent-saturated portion of the filter paper.
- Observe for a color reaction within 30 seconds.
Pos = Violet to purple
Neg = colorless (or light pink/light purple after 30 seconds)
Positive QC: Pseudomonas aeruginosa
Negative QC: Escherichia coli
Extra notes
- Do not read after the 30 second time limit.
- Do not over-saturate the filter paper with reagent.
- Do not use a nichrome or iron-containing loop.
- Colonies should be grown on non-selective, non-differential media and the test works best if they are 18-24 hours old.
- This test is intended for non-fermenters and miscellaneous Gram-negative bacteria.
- Viscid colonies may produce false negative results due to poor penetration of reagent.
- Some organisms, e.g. Pasteurella sp. may be weakly oxidase positive. If a negative result does not match with other biochemical results, repeat the test.
Oxoid™ Oxidative-Fermentative Media (OF Media – Hugh and Leifson) Test Procedure
OF media are used to detect the acid produced by microorganisms that require oxygen for the degradation of carbohydrates. Alternatively, the OF Dextrose (Glucose) medium also allows for the categorization of microorganisms as fermenters, oxidizers or non-utilizers.
The Hugh and Leifson OF test utilize bromthymol blue as the pH indicator.
A. Method for determining if an organism is a fermenter, oxidizer or non-utilizer:
- For each organism being tested, use one (1) OF Control and two (2) Dextrose tubes.
- Pick up an entire isolated colony from an 18-24 Once you have ascertained that all tests worked properly, complete a report for each patient sample including as much about the organism identification as you know.
- Example of reporting: 4+ Candida krusei
- Hand in your worksheets and report forms.
- Pick up an entire isolated colony from an 18-24 Once you have ascertained that all tests worked properly, complete a report for each patient sample including as much about the organism identification as you know.
- hour culture with a sterile straight wire and inoculate the OF Control by stabbing once to approximately ¾ of the way down the tube.
- Next inoculate the two OF Dextrose tubes in the same way by touching colonies and stabbing each approximately ¾ of the way down the tube.
- Overlay one of the Dextrose tubes with a layer of sterile mineral oil (“closed” tube). Loosen the cap of the other tube (“open” tube).
- Incubate the tubes in ambient air at 35-37°C for 24 hours.
- Observe for color changes. Note that the OF Control does not contain carbohydrate so there should be no color change in it. This tube is used for comparison with the carbohydrate-containing tubes. If the color of the control has changed to yellow, do not interpret the rest of the tubes.
- If the Dextrose tubes show no color change at 24 hours, re-incubate and examine daily for up to 4 days.
| Interpretation | Open Tube | Closed Tube |
|---|---|---|
| Fermenter | Acid (yellow) | Acid (yellow) |
| Oxidizer | Acid (yellow near the surface) | Alkaline (green) |
| Non-utilizer | Alkaline (Blue/green) | Alkaline (green) |
Quality Control
| Organism | Basal Control | Dextrose |
|---|---|---|
| Pseudomonas aeruginosa | no change | oxidation |
| Klebsiella pneumoniae | no change | oxidation0/fermentation |
| Alcaligenes faecalis | no change | no change |
Notes
- If the oxidation reaction is delayed or weak, an alkalinity may be observed on the surface but with further incubation, the alkaline reaction reverts to acid.
- Incubation up to 10 days may be necessary.
- Organisms may attack carbohydrates at different rates. Reaction rates for different tubes may therefore vary.
- Microorganisms that can only oxidize glucose (and not ferment it) will not ferment any other carbohydrate. In determining the utilization of other carbohydrates by these microbes, the sealed (“closed”) tube may be omitted as described in Method B below.
B. Method for organisms that require oxygen for the degradation of carbohydrates:
- For each organism being tested, use one (1) OF Control and one (1) each of the specific OF carbohydrate tubes (e.g. lactose, maltose, mannitol, xylose).
- Pick up an entire isolated colony from an 18-24-hour culture with a sterile straight wire and inoculate the OF Control by stabbing once approximately ¾ of the way down the tube.
- Inoculate each carbohydrate-containing tube using the same technique.
- Loosen the lids and incubate the tubes in ambient air at 35-37°C for 24 hours.
- Observe for color changes. Note that the OF Control does not contain carbohydrate so there should be no color change in it. This tube is used for comparison with the carbohydrate-containing tubes. If the color of the control has changed to yellow, do not interpret the rest of the tubes.
- If the carbohydrate-containing tubes show no color change at 24 hours, re-incubate and examine daily for 2-4 days.
Interpretation
Pos = yellow (acid)
Neg = alkaline or no change (blue or green)
| Quality Control | Pos (acid) | Neg (alkaline or no change) |
|---|---|---|
| Dextrose | Pseudomonas aeruginosa | Alcaligenes faecalis |
| Mannitol | Burkholderia cepacia | Stenotrophomonas maltophilia |
| Maltose | Stenotrophomonas maltophilia | Pseudomonas aeruginosa |
| Lactose | Burkholderia cepacia | Pseudomonas aeruginosa |
| Xylose | Pseudomonas aeruginosa | Alcaligenes faecalis |
Remel PathoDx® Streptococcal Latex Grouping Procedure
Note: this latex agglutination procedure is for β-hemolytic streptococci only.
- Allow reagents to come to room temperature.
- Mix reagents thoroughly before use.
- Label a test tube with your organism ID.
- Add 2 drops of Extraction Reagent 1 to the test tube (squeeze the bottle gently by holding vertically over the test tube).
- Add 2 drops of Extraction Reagent 2 to the same test tube.
- Using an applicator stick or inoculating loop, pick 2-4 well-isolated streptococcal colonies from a BA plate (Note: a sweep may be necessary if the colonies are tiny). Emulsify the organisms in the reagents in the test tube by rubbing the stick/loop against the inner bottom or sides of the test tube. Discard the stick (or flame the loop).
- Add 4 drops of Extraction Reagent 3 to the test tube.
- Mix tube by tapping the outside with a finger. Proceed with testing within 60 minutes.
- Label test ovals on the PathoDx® slide with each antiserum being tested. (Be sure to include your suspect grouping as well as a second one as a negative control).
- Using a plastic transfer pipette or a disposable Pasteur pipette, add 1 drop of organism extract to each oval to be used. Discard pipette.
- Mix latex reagents by inversion. Add 1 drop of appropriate antiserum to each oval.
- Mix with a stick, being careful to use a fresh stick for each test oval. Discard sticks.
- Rock or tilt the slide back and forth for up to 60 seconds. Observe for agglutination of latex particles. Record results. Discard slide.
- If both serogroups are negative, you may use the leftover extract to test against other PathoDx® antisera (if available).
Pos = Visible agglutination with background clearing within 1 minute
Neg = No agglutination within 1 minute
QC: The kits are QC’d once per week with known organisms that are appropriate for the antisera being tested (e.g. Streptococcus pyogenes is the positive control for Group A latex but it can also be used as the negative control for Group B, C, F and G latex).
A patient negative control should be done each time you prepare a patient extraction, e.g. if an unknown organism tests positive with group B antisera, the extract must also be tested against a second antiserum (A, C, F or G) to demonstrate that there is no non-specific agglutination taking place.
Phenylalanine Deaminase Test Procedure
Note: This test name may be abbreviated as “PDA”, “PPA”, “PAD” or “PA”
OXOID® Phenylalanine (PAD) Agar Slant
- Touch one or more isolated colonies from an 18-24-hour culture with a sterile loop or stick and inoculate the surface of the slant using a fishtail motion.
- Incubate the tube with a loosened lid in ambient air at 35-37°C for 18-24 hours.
- Add 2-3 drops of 10% ferric chloride directly to the surface of the slant. Be careful not to contaminate the tip of the reagent dropper bottle.
- Observe for immediate formation of a green color on the slant.
Pos = green color
Neg = no color change (note that ferric chloride has a yellow color)
Positive QC: Proteus mirabilis
Negative QC: Escherichia coli
Notes
- The green colour of a positive test fades quickly so an interpretation of positive or negative must be made within 10 minutes of adding the reagent.
- Rolling the ferric chloride over the slant aids in production of a faster reaction and more pronounced color.
- The stability of ferric chloride should be checked weekly.
Phenol Red Sugar Procedure (Carbohydrate Fermentation Test)
OXOID® Phenol Red Broth with 1% Rhamnose
- Pick 1-2 isolated colonies from an 18-24-hour culture with a sterile loop or stick and inoculate the phenol red broth.
- Incubate the tube with a loosened lid in ambient air at 35-37°C for 24-72 hours.
- Examine for a color change.
Pos = yellow (acid)
Neg = red or no change (alkaline)
Delayed reaction = orange (re-incubate)
Positive QC: Escherichia coli
Negative QC: Proteus mirabilis
Notes
- Failure to incubate with a loose lid may produce false negative results.
- Prolonged incubation may be required by some organisms.
- Some microbes may not grow in phenol red medium. Tubes must show growth for the test to be valid.
PGUA (Β-Glucuronidase) Test
Note: This is an alternative test for beta-glucuronidase that is used by some labs in Nova Scotia. We will not be performing it in this lab, but this information is supplied for your future reference.
Principle
Escherichia coli has the ability to break down the substrate p-nitrophenyl-β-D- glucuronide via the enzyme β-glucuronidase.
Procedure
Inoculate a PGUA tube with the unknown organism. Record the time of inoculation on the tube or on your worksheet. Incubate at 35°C. Read at 1 hour. If negative, re-incubate up to 4 hours.
Results: Pos = yellow
Neg = no color
Porphyrin Test Procedure
Oxoid™ and QE II Porphyrin Test Procedure
- Bring the substrate to room temperature before inoculation.
- Use a sterile stick or loop (NOT a swab!) to inoculate the porphyrin (ALA) tube with a heavy suspension of organism taken from chocolate agar. The substrate should appear very cloudy after inoculation.
- Incubate the tube in ambient air at 35-37oC for 18-24 hours.
- Examine the tube in a dark room or cupboard with a Wood’s lamp (ultraviolet lamp). A red to orange fluorescence indicates the presence of porphyrins and the test is interpreted as positive.
- Add an equal volume of Kovac’s reagent to the tube and mix well. Allow the substrate and reagent to separate. Observe for a red or pink colour in the lower layer which indicates the presence of porphobilinogen.
Interpretation
Pos: the formation of a red (pink) color in the lower aqueous phase indicates porphobilinogen. Alternately, fluorescence under a Wood’s lamp indicates the presence of porphyrins. This organism does not require an extraneous source of X factor since it can make its own.
Neg: No colour after addition of Kovac’s. No fluorescence under a Wood’s lamp.
Quality Control
Positive QC: Haemophilus parainfluenzae ATCC 7901 Negative QC: Haemophilus influenzae ATCC 10211 or 43065
Notes
- The porphyrin test is used to differentiate species of Haemophilus which require an external source of X factor from those that do not. It detects the presence of porphobilinogen synthase which converts δ-aminolevulinic acid (ALA) to heme (X factor). Porphobilinogen and porphyrins are products of this reaction.
- A false negative reaction may occur if the inoculum is too light or if the colonies being tested are more than 24 hours old.
- It is very important to interpret the results carefully after addition of Kovac’s reagent. The color change being observed is in the aqueous portion (the lower phase). If a pink color is observed in the upper layer (alcohol phase), it is indicative of the presence of indole. A porphyrin tube that shows pink in the top layer and clear in the bottom layer is an indole-positive organism that cannot produce its own X factor.
PYR Test Procedure
Remel™ PYR Test Procedure
- Use forceps to place a PYR disk on a clean microscope slide.
- Moisten the disk slightly with sterile water. DO NOT over-saturate the disk (the disk should not be flooded/floating).
- Use a sterile stick or loop to pick up several isolated colonies (4-10) from an 18-24 hour culture.
- Rub the inoculum gently onto a small area of the disk.
- Incubate at room temperature for 2 minutes.
- Add one (1) drop of color developer (dimethylaminocinnamaldehyde) to the disk.
- Allow up to 1 minute for color development.
Pos = Pink to red color within 1 minute of adding color developer
Neg = Cream, yellow, or no color change within 1 minute of adding color developer.
Positive QC: Streptococcus pyogenes, Enterococcus sp. Negative QC: Streptococcus agalactiae
6.5% Salt Tolerance Test Procedure
Oxoid™ Salt Tolerance Broth 6.5% (6.5% NaCl Test)
- Use a sterile stick or loop to lightly inoculate the broth medium with 1-2 colonies.
- Incubate the tube with a loosened lid in ambient air at 35-37°C for 24 hours.
- Observe the tube for turbidity. Negative reactions can be re-incubated up to 72 hours.
Pos = growth (turbidity)
Neg = no growth (clear)
Positive QC: Enterococcus sp.
Negative QC: Streptococcus bovis
Spore Stain
- Prepare a smear of the unknown. Allow to air dry and heat-fix.
- Place a small piece of bibulous paper or filter paper over the smear and saturate it with malachite green.
- Heat the slide gently over a Bunsen burner for 5 minutes. Be sure to keep the bibulous paper damp with malachite green.
- Remove the bibulous paper and rinse the slide gently with water.
- Counterstain with safranin for 2 minutes.
- Rinse gently with water.
- Blot dry.
- Observe microscopically for presence and location of spores.
Interpretation
Endospores: green
Vegetative cell: pink
Quality Control
Bacillus spp.: pos
Corynebacterium spp.: neg
Notes
- It is important that the bibulous paper remains moist so that the dye is steamed into the bacterial cell and not baked.
Starch Gelatin Carbohydrate Media (Carbohydrate Utilization Test for Fastidious Bacteria)
Carbohydrate oxidation test media used for the identification of Neisseria spp.
- Use a sterile loop to remove a full loopful of growth from pure identical colonies.
- Heavily inoculate the starch gelatin control tube and the specific starch gelatin carbohydrate tube(s) by making several shallow stabs into the upper ¼ inch portion of each tube.
- Incubate the tube with a loosened lid in ambient air at 35-37°C for 18- 48 hours.
Interpretation
Pos = yellow
Neg = no color change
Quality Control
| Dextrose | Maltose | Lactose | Sucrose | |
|---|---|---|---|---|
| N. gonorrhoeae | + | – | – | – |
| N. meningitidis | + | + | – | – |
| N. lactamica | + | + | + | – |
| N. sicca | + | + | – | + |
| M. catarrhalis | – | – | – | – |
Notes
- Compare the specific carbohydrate tube with the control tube which does not contain carbohydrate. Since the carbohydrate is used oxidatively and only a small amount of acid may be formed, only a slight change may be detected.
- A deep red color indicates peptone utilization and a lack of carbohydrate utilization.
- Lack of sufficient inoculum or use of old cultures may result in no degradation of the carbohydrate.
- Incubation in carbon dioxide will produce a yellow color change unrelated to carbohydrate utilization.
- Inoculation to the bottom of the tube may make a weak acid reaction difficult to read.
- Failure to incubate with a loose lid will produce no color change or a weak one.
- Negative tests can be re-incubated up to 48 hours.
Starch Hydrolysis Test Procedure – Ivany
- Use a sterile loop to inoculate organisms from an 18-24-hour culture with a heavy single streak to the surface of a Mueller Hinton agar plate.
- Incubate at 35-37°C in ambient air for 48 hours.
- Flood the plate with Gram’s iodine.
Pos = a clear zone surrounding the organism growth
Neg = dark blue, purple or black color up to the edge of the organism growth
Positive QC: Streptococcus bovis
Negative QC: Enterococcus sp.
Triple Sugar Iron Agar Test Procedure
OXOID® Triple Sugar Iron Agar (TSI)
- Use a sterile straight wire to inoculate a single colony from a pure 18-24-hour culture.
- While inoculating, stab the center of the agar to the base of the butt and then streak the slant with a fishtail motion. Be careful not to disturb the integrity of the medium.
- Incubate the tube with a loosened cap in ambient air at 35-37°C for 18-24 hours.
Interpretation
| Alk/acid (red slant/yellow butt) | Glucose fermentation only |
|---|---|
| Acid/acid (yellow slant/yellow butt) | Glucose and one, or both, of lactose/sucrose fermentation |
| Alk/NC or Alk/alk (red slant, red butt) | No carbohydrate fermentation. Aerobic organism |
| Blackening of medium: | H2S produced |
| Bubbles, splits, cracks or pushing up of agar: | Gas produced |
Quality Control
| Salmonella typhimurium | Alkaline slant, H2S, acid butt, gas |
| Escherichia coli | Acid slant, no H2S, acid butt, gas |
| Shigella flexneri | Alkaline slant, no H2S, alkaline butt, no gas |
| Pseudomonas aeruginosa | Alkaline slant, no H2S, no change in butt (aerobic) |
Notes
- Failure to incubate with caps loose may produce erroneous results.
- TSI must be read within 18-24 hours. If read earlier, a false positive reaction may occur. If read later, a false negative reaction may occur.
- It is important to stab the butt of the agar: failure to do so will make the test invalid. Be careful to avoid mechanical splitting of the agar while stabbing the butt. This may give a false appearance of gas production.
- An H2S-producing organism may mask the acid production in the butt. Since H2S production requires an acid environment, assume that the butt portion is acid if H2S is present.
Urea Test Procedure
OXOID® Urea Agar
- Touch one or more isolated colonies from an 18-24-hour culture with a sterile loop, straight wire or stick and inoculate the entire surface of the slant using a fishtail motion.
- Incubate the tube with a loosened lid in ambient air at 35-37°C for 4 hours. Examine for a color change and re-incubate if negative. Examine again at 18 hours. Re-incubate up to 48 hours if still negative.
Pos = Pink-purple color on slant and/or butt
Neg = no color change from pale orange color of medium
Positive QC: Proteus mirabilis
Negative QC: Escherichia coli
Notes
- Failure to incubate with the cap loose may lead to erroneous results.
- Proteus spp. will split urea soon after inoculation. Positive results can therefore be read within the first 3-5 hours after incubation (this is considered to be a rapid positive result).
- Some Enterobacteriaceae (e.g.Citrobacter freundii, Klebsiella pneumoniae) may take up to 48 hours to split urea (this is considered to be a delayed positive or weak positive result).
Voges-Proskauer (VP) Procedure OXOID® Voges-Proskauer (VP) Media
- Touch an isolated colony from an 18-24-hour culture with a sterile loop, straight wire or stick and inoculate the VP broth. (Note: if the colonies are small, use two).
- Incubate the tube with a loosened lid in ambient air at 35-37°C for 48-72 hours.
- Mix the contents of the VP broth. Remove about 1 mL of broth into a 16×100 mm test tube. First add 6 drops of alpha naphthol and then 2 drops of 40% KOH to the aliquot.
- Carefully shake the tube for 30 seconds to 1 minute to expose the medium to atmospheric oxygen.
- Incubate 30-60 minutes at room temperature.
- Observe for color development.
- If the test result is negative, re-incubate the original broth and re-test daily for up to 5 days.
Pos = a red color at the broth surface (or throughout)
Neg = no color at broth surface
Positive QC: Enterobacter cloacae
Negative QC: Escherichia coli
Notes
- The order in which the VP reagents are added is important. The alpha-naphthol is added first! A reversal in the order of the reagents may give a weak positive or false negative result.
- Shaking the tube after the addition of reagents is necessary to ensure the oxidation of acetoin, which results in the color change
- A copper-like color after the addition of alpha-naphthol is interpreted as negative.
- Excess KOH may mask a weak positive reaction.
- Delayed reading (past 1 hour) may give a false positive result.
- Because some organisms destroy acetoin, VP test results should always be interpreted in context with indole and citrate results.
- Occasionally, a known VP -positive organism may not give a positive VP reaction. According to some methods, the aliquot with added reagents can be gently heated (e.g. in the incubator) for up to 1 hour. A VP-positive organism should produce a positive result after heating. Incubation after one hour, however, could result in false positive results. Check your manufacturer’s guidelines to ascertain if this modification is reliable.
Wet Motility Test
Gently emulsify a small amount of a colony in a fairly large drop of saline on a labeled microscope slide. Place a coverslip gently on top. If bubbles are present, quickly pass the preparation over a Bunsen burner flame or in front of a micro incinerator element to drive them out. Do not allow the preparation to stand as it will dry up if not examined within 10 to 15 minutes.
Observe the wet preparation on the microscope by examining under 10X and 40X objectives but NOT oil immersion. (Be very careful. This preparation contains live organism and the coverslip is floating loosely on the surface. Tipping the slide or moving to the 100X objective will cause the fluid to seep or ooze from the edges of the coverslip and possibly contaminate the microscope.) Check for directed movement as opposed to Brownian movement. Discard the slide in biohazard garbage.
Note: since no stain has been applied, this preparation is colorless. Any color you may see is due to chromatic aberration.
Alternative procedure: Inoculate a colony to a sterile broth and incubate at room temperature for 20-30 minutes. Prepare a wet prep and observe for motility.
X and V Factor Test Procedure
Oxoid™ X, V and XV Factor Discs
- Bring a Mueller Hinton (or a trypticase soy plate with NO blood) plate and discs to room temperature.
- Prepare a light saline suspension equivalent to a 0.5 McFarland standard of the organism to be tested. It is important to use a sterile loop, rather than a swab, to avoid X-factor carryover from the medium that the organism is taken from.
- Dip a fresh sterile cotton swab into the well-mixed suspension. Roll the swab evenly over the entire surface of the plate in three planes.
- Use the same swab to inoculate a chocolate agar plate (labeled with the specimen identification and the words “purity plate”) and then discard the swab into a biological waste container. Use a sterile loop to streak the purity plate for isolated colonies.
- Aseptically place X, V and XV discs on the inoculated agar plate spacing them evenly and at least 10-20 mm from the edge of the plate. Do not move the discs once they have touched the agar surface.
- Incubate the plates at 35 ± 2° C in a CO2 incubator or candle jar for 18-24 hours. If necessary, the X, V, XV plate can be re-incubated up to 48 hours.
- After incubation, evaluate the purity plate to ensure that the organism suspension used for the test was pure (if done).
- Examine for growth at the edge of each disc.
Interpretation
- Growth around XV disc only: organism requires an extraneous source of both
- Growth around the V disc, no growth around the X disc and light growth around the XV disc: organism requires an extraneous source of V factor (can make its own X).
- Growth around the X disc, no growth around the V disc and growth around the XV disc: organism requires an extraneous source of X factor (can make its own V).
- Growth over entire surface of agar – no requirement for X or V
Quality Control/Examples:
| X Factor | V Factor | X+V Factor | |
|---|---|---|---|
| Haemophilus influenzae | no growth | no growth | growth |
| Haemophilus parainfluenzae | no growth | growth | growth |
| Haemophilus ducreyi | growth | no growth | growth |
| Haemophilus aegyptius | no growth | no growth | growth |
| Aggregatibacter aphrophilus | growth | growth | growth |
| Bordetella pertussis | growth | growth | growth |
Notes
- Plate may be re-incubated if there is no growth around any discs at 24 hours.
